UPDATE: A groundbreaking study published on July 5, 2025, by researchers at KU Leuven, has unveiled a revolutionary deep learning model that accurately analyzes the 3D microstructure of fruit tissues, surpassing traditional 2D methods. This innovative approach promises to significantly enhance the understanding of plant physiology and storage behavior in apples and pears, with immediate implications for agricultural practices worldwide.
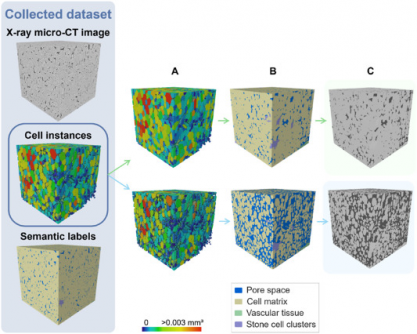
The model, developed by Pieter Verboven and his team, utilizes X-ray micro-CT imaging technology to provide non-destructive, high-resolution insights into the complex architecture of fruit tissues. Previous methods struggled with low image contrast and sample preparation issues, but this new framework offers a comprehensive solution, paving the way for faster, more precise studies.
In a remarkable achievement, the 3D model reached an Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, outperforming both a 2D instance segmentation model and traditional algorithms. This leap in accuracy is critical for researchers seeking to explore how microscopic structures impact water, gas, and nutrient transport in fruits.
The deep learning framework employs a sophisticated 3D panoptic segmentation method, enabling the identification of individual parenchyma cells and various tissue types, including vascular tissues and stone cell clusters. This automation not only reduces manual labor but also enhances the precision of tissue characterization.
In practical terms, this development could transform fruit research by revealing how cellular arrangements determine texture, storability, and susceptibility to physiological disorders such as browning or watercore. The implications extend beyond fruit, as the technology is designed to be scalable for studying various crops, influencing the future of agricultural science.
Preliminary visual validations confirmed the model’s accuracy, successfully identifying vascular bundles in Kizuri and Braeburn apples, and providing realistic segmentations of stone cell clusters in Celina and Fred pears, achieving a Dice Similarity Coefficient (DSC) of up to 0.90.
As researchers face the challenges of dataset imbalance and domain shifts, the study emphasizes the need for further augmentation techniques. However, with its compatibility with standard X-ray micro-CT instruments, this deep learning model stands out as an accessible, powerful tool for integrating artificial intelligence into plant anatomy and food science research.
The implications of this study are vast, offering a new paradigm for understanding plant tissue microstructure. As the agricultural sector seeks to adapt to changing environmental conditions and consumer demands, this research could accelerate the development of crops that are more resilient and better suited for storage.
Researchers and agricultural professionals alike are urged to explore this new technology, which could significantly enhance the efficiency and accuracy of plant studies. This development is a major step towards revolutionizing how we study and interact with the biological structures that underpin our food systems.
Stay tuned for more updates as this research unfolds and its applications expand across the agricultural landscape.